Featured Story
![]()
March 2007
Bar Coding for Botany
![]()
A system modeled on commercial bar codes may soon enable
anyone to identify any plant from a small fragment of its DNA.
![]()
By Kenneth M. Cameron
|
||
What the heck are these? The documents for this crate say the contents are Polypodium ferns. Those are perfectly legal to import, but all the leaves have been hacked off these plants. I can’t identify them from the stems alone. Jim, can you get a reading on them?”
“Sure—just a second. . . . Well, according to my Global Flora Scanner, they’re actually Stangeria eriopus, the Natal grass cycad, which looks a lot like a fern. It’s an endangered species from Mozambique—says here they’re just about extinct in the wild. They’re illegal to import, but collectors are just crazy about them. Apparently some cycads sell for as much as $20,000 on the black market. I’ve never intercepted Stangerias here at the airport before. Good thing you spotted them—and that they were in the GFS database. We’d better investigate; this should mean a big fine or even an arrest for the importer.”
The dialogue might sound like science fiction, but that kind of scenario could transpire sooner than you think. One of the great biological projects of our time will be to collect DNA sequences from every living species on Earth. The objective is to create a universal genetic database of life. Once it is mostly complete—perhaps a decade from now—the project will enable any plant, animal, fungus, or other organism to be identified simply by sampling its DNA and comparing that with the database of known DNA sequences.
That comprehensive approach to identifying species is called DNA bar coding. As the name implies, the idea is to develop, as explicitly as possible, the analogy with the universal product codes, or bar-code labels, that are attached to nearly every consumer product, from applesauce to zucchini bread. What makes the analogy such a good one?
|
||
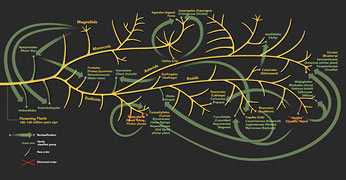
DNA bar coding is the newest of several techniques that promise to make important contributions to the basic science of systematic biology. The discipline seeks to identify and classify organisms, reconstruct their evolutionary history, and map the extent of biological diversity—in other words, to build the family tree of life. The use of molecular tools in pursuing those goals has already transformed the way biologists understand the natural world. In particular, the wide availability of DNA bar coding in the future could enable specialists to make rapid, reliable identifications in the field, and make it possible for armies of amateur naturalists to contribute to the study of the range and diversity of species. Within botanical circles, the influence of molecular data on systematics has been revolutionizing the study of plants in the laboratory and in the field.
Since plant systematists first began comparing gene sequences in the 1980s, their studies, more often than not, have simply confirmed classifications that botanists have accepted for centuries. For example, molecular evidence confirms that almonds, apples, cherries, pears, and strawberries are all closely related; all of them are best classified with roses in a plant family called the Rosaceae.
|
||
But nearly every study in molecular systematics has also led to its share of surprises. More than ten years ago, DNA data showed that, contrary to the accepted thinking of the day, a number of carnivorous plants that employ radically different methods of capturing animals share a common ancestor. A molecular phylogenetic tree showed that Old World pitcher plants of the genus Nepenthes are closely related to sundews (Drosera) and to Venus flytraps (Dionaea muscipula), even though the three plants evolved three distinct ways of catching prey: fluid-filled pitfall traps, sticky flypaper traps, and rapidly closing snap traps.
More recently, my collaborators and I demonstrated that Aldrovanda vesiculosa, the carnivorous waterwheel plant, is also a member of that highly unusual group. Like the Venus flytrap, Aldrovanda catches its dinner in snap traps. But unlike all other members of the group, it is aquatic. As if that finding were not strange enough, our studies also showed that the same carnivorous-plant group is related to buckwheat, cactus, carnation, jojoba, rhubarb, and salt cedar. Today botanists classify all of them in distinct but closely related families of the plant order Caryophyllales.
Perhaps the most dramatic example of a revised classification brought about by molecular systematics is Nelumbo, the water lotus. Cultivated for its beautiful flowers, distinctive seedpods, and edible underwater rhizomes, the lotus has been immortalized in Chinese paintings for centuries. Most people, including botanists, assumed it must be related to water lilies or to some other aquatic flowering plant. In fact, according to DNA-sequence data, the lotus is most closely related to Platanus, the sycamore or plane tree, along with the trees and shrubs in the family Proteaceae, which includes macadamia nuts and the showy-flowered members of the genus Protea.
|
||
Superficially, the plants have nothing in common. But when the molecular evidence suggested taking a closer look, botanists discovered that lotuses, proteas, and sycamores share similar floral and vegetative features. Moreover, the group was widespread during the Cretaceous period and probably more diverse in form than it is today, suggesting that plants intermediary among the sycamore, lotus, and protea might once have existed. Examples of surprising relationships revealed by recent molecular analyses go on and on, and include the close kinship of fungi to animals, cucumbers and begonias to oaks, orchids to asparagus, and violets to poinsettias, among many other remarkable glimpses into botanical genealogy.
My sci-fi story of a customs bust is a good way to understand how all that botanical detective work may one day pay off. Trade in a number of plants has been banned or restricted under the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). Among them are species of cactus, cycad, ginseng, orchid, palm, and tree fern. Full-grown adult specimens of those plants are usually easy for customs inspectors to spot during a search. But to thwart the inspectors, smugglers have been known to chop off the plants’ leaves, then illegally import the bare stems under false names. The plants remain alive, of course, and will produce new leaves the following season, but the practice makes it nearly impossible for officials to correctly identify the plants or to take legal action.
| ||
Identifying a plant from its DNA has several important advantages. First, the DNA of each species is distinct from any other; DNA is a unique identifier. Second, all nonreproductive cells of a given organism have the same complement of DNA. Testing any fragment of the organism—whether leaf, root, stem, or petal—is enough to identify the organism. Third, the DNA in each cell of the organism remains unchanged no matter what the current stage in the organism’s life cycle—whether it be a plant in seedling or adult stage, a frog in larval or adult form, or a fungus in hyphal or mushroom phase.
The advantages of the bar-code project are even more pronounced. The bar code itself would presumably be just a unique indexing feature, one diagnostic part, of each cell’s entire complement of DNA. A database of DNA bar codes for all species, if it were available, would simplify a customs inspector’s job. He or she would sample a few cells of virtually any plant or plant fragment that came through the inspection station. The inspector’s handheld scanner would then sequence the bar-code DNA, submit the bar code for comparison with the universal database online, assign the correct name to the plant material, and link to useful information about the species.
|
||
But the practical applications of DNA bar coding for plants are hardly limited to catching smugglers. I have developed a genetic test to distinguish the vanilla beans of various species. The beans look similar, but they are quite different in quality. Inferior species are occasionally sold—either fraudulently or mistakenly—as premium-quality species to manufacturers of vanilla extracts, a problem DNA bar coding will help eliminate. Consumers will be glad to hear that dried roots, leaves, and stems from medicinal plants can be identified with DNA bar coding before being sold as herbal supplements. Ecologists, too, will find the technique valuable in field surveys, because they will be able to include all plants in an area—whether big or small, easy or hard to identify.
Two other, more universal advantages of DNA bar coding are worth mentioning: it could extend the reach of expertise and make sophisticated biological knowledge more accessible to everyone. It has been estimated that biologists may have discovered and cataloged no more than 10 percent of Earth’s biota. Yet species are disappearing at an alarming rate. That leaves a lot of work to be done in a very short time by taxonomists. Yet taxonomy is a shrinking profession because of budget cuts at museums and academic institutions, and a trend away from organismal biology toward the study of life at the cellular and molecular levels. Moreover, the same taxonomists are asked all too often to devote substantial time and expertise to making routine identifications of well-known species.
With DNA bar coding, any organism could be identified by entry-level technicians. Experts could give up the time-consuming burden of making routine “dets,” or determinations, and focus their energies instead on more substantial scientific tasks. No longer would just a few authorities
|
||
So how far along is the scientific community in developing DNA bar-code databases? In zoology great progress has already been made. A single gene known as cox1, which occurs in the mitochondrial genome, has been chosen as the universal genetic bar code for animals: nearly every animal species possesses a distinct version of cox1. Zoologists in laboratories around the world are sharing techniques for sequencing the gene, and are quickly amassing enormous numbers of cox1 gene sequences from thousands of different species.
One of the best-publicized projects is the All Birds Barcoding Initiative, whose goal is to establish an archive of DNA bar codes for the approximately 10,000 known species of birds on Earth by 2010. Even more ambitious is FISH-BOL, aka the Fish Barcode of Life Initiative, which has already started to collect DNA bar codes for the world’s more than 29,000 known fish species. FISH-BOL hopes to complete its collection within the next five years.
Unfortunately, the botanical community has not been as quick to jump into DNA bar coding as zoologists have. In part, the reason is that plants present unique challenges. Pressed and dried plant specimens in herbaria often yield their DNA less readily than do preserved animal specimens in museums. Moreover, animal species are most commonly defined by their reproductive isolation from one another, whereas many plant species can hybridize, thereby blurring their genetic boundaries. Finally, the mitochondrial genome has evolved quite differently in plants than it has in animals. The cox1 gene is not practical as a universal bar-code marker for photosynthetic organisms.
|
|||
![]()
To address those problems, the Consortium for the Barcode of Life, a body of scientists representing natural-history museums, universities, and botanical gardens around the world, formed a plant working group in 2005. That group, on which I serve as vice-chair, is actively engaged in a two-phase project to find a plant gene, or small set of genes, comparable to cox1 in animals, that can act as a bar code for all plant life. The first phase, completed in early 2006, aimed to identify five or more candidate gene regions from a small set of plants. The second phase is devoted to testing those candidates across the entire plant kingdom. There is consensus among the two dozen scientists in the group that the gene or genes should meet several criteria. The genes should be present in all plants, easy to sequence, as short as possible, and highly variable from plant to plant.
After several months of testing during our first phase, we identified six candidate genes by comparing gene sequences from 122 plants, representing sixty-one closely related species pairs from across the entire plant tree of life. The candidates are all chloroplast genes—known as accD, matK, ndhJ, rpoB, rpoC1, and YCF5—most of which happen to code for proteins that play a role in photosynthesis. Now, in the second phase of the project, those six candidate genes are being sequenced in a broader selection of plants, including various conifers, cycads, ferns, and mosses, as well as monocot and dicot flowering plants. For example, in my laboratory we are sequencing the candidate genes for almost every species of the tropical fern genus Elaphoglossum, the conifer genus Cupressus, and the Hawaiian flowering plant genus Labordia.
My laboratory is also overseeing a complementary project to sequence the six candidate genes in all the vascular plant species of a fixed geographic region, rather than in scattered lineages throughout the entire plant kingdom. And where better to start than in one’s own backyard? Our goal is to bar-code every species of vascular plant—both native and exotic—within the fifty-acre forest at the New York Botanical Garden.
|
||
Many other such projects are taking place around the world, and as a whole the plant working group is making excellent progress. Within the coming months we expect to announce our recommendation for the gene or genes that will enable plant DNA bar coding to proceed.
In spite of the rapid gains DNA bar coding is making, support for it in the biological community is not unanimous. The strongest objection is that the technique is not foolproof. In practice, though, as technology develops, reliability should improve dramatically, and the problem should largely go away.
Modern biology is built around two primary paradigms. One centers on evolution and embraces the disciplines of Mendelian genetics, natural history, and systematics. The merger of those disciplines during the first half of the 1900s was called the modern synthesis. The second paradigm centers on gene expression and is the foundation of biochemistry, cell biology, molecular biology, and physiology. Those areas of research were brought together during the second half of the twentieth century under the unifying framework provided by the structure of DNA; that paradigm has been termed the molecular synthesis.
Biology is on the verge of a great new scientific revolution that will unite those two separate paradigms into a single program of research: the final synthesis. That new paradigm will enable molecular biologists in their laboratories and organismal biologists in the field to begin to communicate across disciplines.
Within botany, the blending of disciplines is already well underway. The molecular revolution has forced botanists to look more carefully at plants as well-known as the Venus flytrap and the water lotus. DNA bar coding promises further progress by providing new tools for scientists, amateur naturalists, and the public at large. Our children, armed with handheld Global Flora Scanners made possible by the molecular studies in full blossom today, will undoubtedly see and hopefully respect the diversity of life on this planet in ways that none of us can now imagine.
 |
Copyright © Natural History Magazine, Inc., 2007